| 2. Biocatalysis |
Enzymes are the workhorses in biological systems. Besides, enzymes are becoming to play key catalytic roles in biological manufacturing because of their high level of efficiency and selectivity, as well as their mild operating conditions in biocatalysis. However, the cost-effective use of enzymes in vitro requires them to function in alternative solvents and/or over wider ranges of temperature and pressure. Therefore, most enzymes need to be engineered or manipulated to satisfy their uses in vitro. To date, protein engineering and directed evolution methodologies have not only greatly improved the characteristics of enzymes for various in vitro applications, but also expanded the range of reactions that enzymes can catalyze, even the reactions that do not happen in nature. These make enzymes possess promising applications in bioproductions, solving environmental problems, offering clean energy and therapeutic development. It is expected that biocatalysis will see a new wave of expansion in the time of rapid artificial intelligence development.
The research on biocatalysis in the lab includes four aspects, that is, rational design and evolution of enzymes based computations and virtual screening including molecular simulations and artificial intelligence, fabrication of multi-enzyme systems, exploration of artificial biocatalytic photosynthesis, and development of new methods and supports for enzyme modifications/immobilizations. |
| 2.1. Computation-based enzyme design and evolutions |
The variety and complexity of enzymatic functions found in nature are astounding. Enzyme promiscuity is especially impressive. For example, >25,000 members of the amidohydrolase superfamily can catalyze >40 unique reactions. Enzyme promiscuity also provided opportunities for enzymes to evolve. Many enzymes have recently evolved for the degradation of xenobiotic compounds (pesticides, explosives) and antibiotics, which were synthesized just several decades ago.
It is a long time for enzyme to gain new function through natural evolution. In order to satisfy the demand for biocatalysis, many efforts have been devoted to redesign enzymes. Directed evolution has now become a common method that can enhance natural evolution a million fold, which is successfully used in improving catalytic efficiencies and specificities of many enzymes. Although negative results generally remain unpublished, many directed evolution experiments come to an early halt after only a minor improvement or no improvement at all.
In recent years, rational design based on in silico protein design has become widely used in enzyme design with the development of computational technology and protein structure analysis. Compared to directed evolution or site-directed mutagenesis, rational design could break through the limits of natural evolution and greatly reduce the amount of experiment to obtain more rapid protein evolution. The basic principle of rational design is that protein structure dictates its function. The protocol of rational design of enzymes is shown in Figure 1.The rational design of enzymes can start with natural enzymes or de novo designed enzymes as the backbone. Then multiscale molecular dynamics (MD) simulations can be performed to obtain information for mutating enzymes. The mutations are ranked based on the energy of structure to select stable structures. These structures are evaluated using experimental validation to select the optimized enzyme that fit the desired protein function.
Rational design of enzymes can realize the conversion of enzyme function, which is a promising and expanding research field. We devote to increase enzyme activity towards specific substrates, improving thermal stability of enzymes and changing substrate preference of enzymes through rational design of enzymes. |
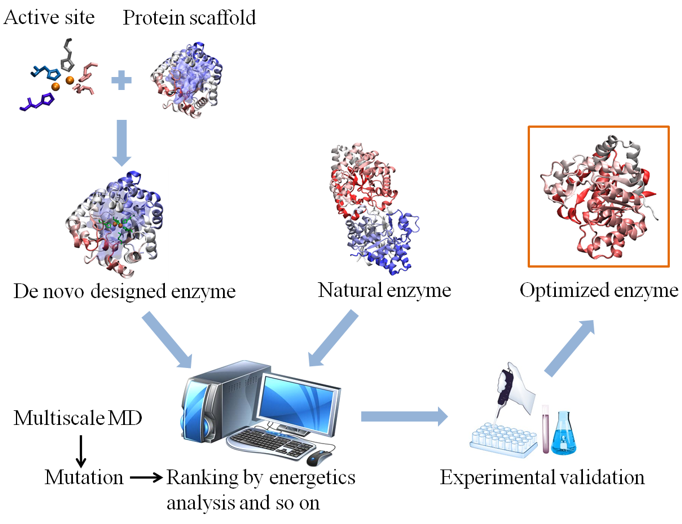 |
Figure 1. Schematic for the protocol of computation-based design/redesign of enzymes. |
| 2.2. Multi-enzyme systems |
Recently, in vitro biocatalytic systems are considered as a potential alternative to traditional chemical synthesis. Also, the development of systems biology and synthetic biology provides convenience to obtain efficient biocatalysts. On this basis, we can design more efficient multi-enzyme cascade reaction to get the high-value compounds in a greener and more efficient way.
In organisms, complex chemical transformations are common and usually involve two or more enzymatic reactions that are linked by different steps (sequential, coupled, divergent, convergent). And the enzymes required for the multi-enzyme cascade reactions are often restricted to specific regions or to different clusters in microorganisms. This specific arrangement leads to more stable structures and facilitates substrate channeling between enzymes, thus improving the catalytic reaction rate and product yield. Inspired by nature, researchers have used a variety of ways to achieve multi-enzyme reactions in vitro, including enzyme fusion, multi-enzyme self-assembly and multi-enzyme immobilization (Figure 2).
Oxidoreductase is the largest class of enzymes, and a number of bioconversions involve oxidation/reduction reactions. Therefore, oxidoreductase plays an important role in the production of organic acids, amino acids, steroids and chiral drugs, etc. Coenzymes are required in oxidoreductase catalyzed reactions, which act as hydrogen or electron transporters in the reaction process. Among the coenzymes, nicotinamide coenzymes NAD(H) and NADP(H) are the main coenzymes of oxidoreductase. However, the large-scale application of oxidoreductase in industry is greatly limited by the nature of coenzymes, including high costs, unstability, non-recycling and regeneration after reaction. To overcome the drawbacks, it requires the development of efficient and economical regeneration techniques for multiple recycled utilization of coenzymes.
Coenzyme regeneration can be categorized into enzymatic, chemical, electrochemical and photochemical methods. Of those, the enzymatic regeneration of coenzymes has the advantages of mild reaction conditions, green chemistry, high total turnover number (TTN) and high selectivity, so it is a good choice for coenzyme regeneration (Figure 3).
In order to achieve high efficiency and sustainability of enzymatic reaction, we are focusing on the development of new self-assembly systems for multi-enzyme cascade reactions in vitro and efficient recycling and regeneration systems for the nicotinamide coenzymes. |
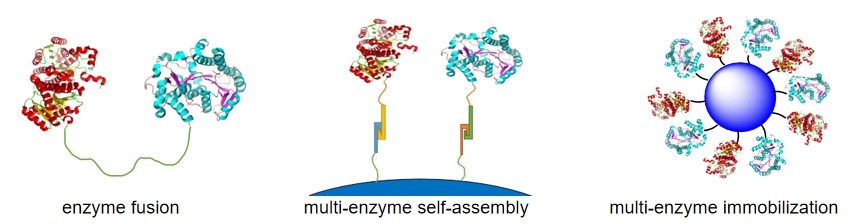 |
Figure 2. A schematic illustration of multi-enzyme reaction strategies in vitro. |
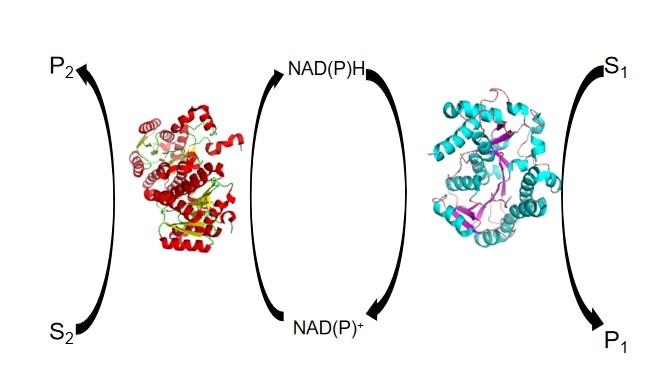 |
Figure 3. A schematic illustration of the enzymatic approach for coenzyme regeneration. |
| 2.3. Artificial biocatalytic photosynthesis |
With the increasing demand of traditional fossil resources in the whole world, solar energy has drawn a tremendous interest of researchers as a clean, cheap, and sustainable energy source. Therefore, the enhanced efficient methods for solar energy conversion are a challenging and promising field nowadays.
An important way for solar energy conversion is to imitate the natural conversion pathways. As a fundamental chemical reaction system in nature, the photosynthesis system exists in green plants, algae and bacteria, and converts the light energy into stable chemical energy in energy-rich compounds. For the photosynthesis system in the thylakoids of green plants, the photosynthesis reaction consists of photoreaction and dark reaction. The photoreaction refers to the conversion of solar energy into the chemical energy in ATP and coenzyme NADPH under illumination, while the dark reaction, namely Calvin Circle, fixes free CO2 into carbohydrates using the emerging ATP and NADPH.
To construct effective artificial photosynthesis systems, the reaction mechanism of photosynthesis ought to be understood. Generally, the photoreaction carries on in three steps, i.e., solar energy capture, photo-induced charge separation and electron transfer, and synthesis of NADPH and ATP. Under illumination, the light is absorbed by antenna complexes, then the energy converges at the primary photosensitive donor P680, inducing the primary charge separation. After photo-induction, the electrons transfer from P680 to ferredoxin-NADP-reductase and ATP synthase by a series of protein-cytochrome complexes, to participate in the synthesis of NADPH and ATP. Meanwhile, water molecules around P680 acting as final electron donors are oxidized into protons and oxygen molecules, providing electrons for the regeneration of P680. For artificial photosynthesis systems, as shown in Figure 4, there are also three functional parts for energy conversion, i.e., photosensitizer, redox enzyme system and electron donor agent, for light-harvesting, catalyzing redox reaction and photosensitizer regeneration, respectively.
Therefore, electron transport is a vital process for light-driving redox reactions, and the natural oxidoreductases often rely on a cofactor for the conversion of light energy and the electron transport from photosensitizer to redox product. As mentioned above, over 80% of oxidoreductases depend on the nicotinamide adenine dinucleotide cofactors, NAD(P)H, which are generally too expensive to be employed in industry. Our research thus focus on the regeneration of NAD(P)H through biocatalytic methods in photosynthesis systems, or even surmounting the dependence of NAD(P)H. |
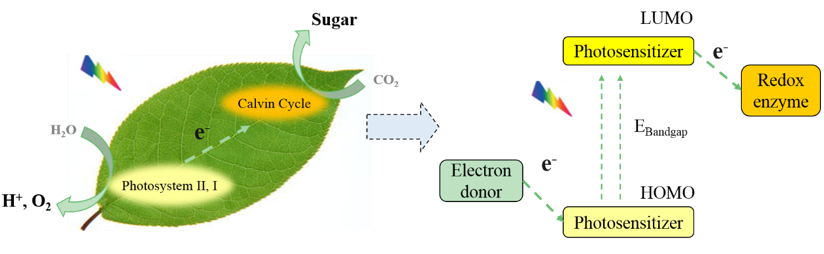 |
Figure 4. Schematic illustration of (left) natural photosynthesis and (right) artificial biocatalytic photosynthesis. |
| 2.4. Enzyme modifications/immobilizations |
The process of enzyme-based biocatalyst has been greatly used in various fields of biotechnology due to its high substrate specificity, ease of production and environmental friendliness. However, the practical applications of natural enzymes are often hampered by several drawbacks including high costs, low stability and difficulty in recycled use. Enzyme modifications and immobilization are utilized to enhance the stability and reusability of free enzymes, making them industrially and commercially viable. Immobilization means confining enzyme molecules to a certain space or completely attaching them to a solid structure, to retain in a proper geometry. The performance of immobilized enzymes depends on support materials and immobilization methods.
Figure 5 illustrates a schematic overview of support materials and methods used for immobilization purposes. The traditional and most commonly used immobilization methods can be categorized into four types, that is, adsorption, covalent attachment, entrapment, and cross-linking. Each has its own advantages and disadvantages. However, there is no universal method to immobilize enzyme molecules. All immobilization approaches require an extra support material except the cross-linking method. Various materials have been used as carrier matrices, usually inert polymers and inorganic materials. Our research has focused on development of biocompatible zwitterionic supports that afford three-dimensional spatial immobilization of enzyme molecules for enhanced enzyme activity and/or stability.
Recently, it has been found that zwitterionic supports such as poly(carboxybetaine methacrylate)-grafted silica nanoparticle (SNPs-pCBMA) are beneficial to the stabilization of enzymes immobilized on it. The mechanism of improved enzyme stability is considered due to protective effects of zwitterionic polymer on stabilizing protein structures. Namely, zwitterionic polymer can create a superhydrophilic microenvironment around the immobilized proteins, weakening water mobility, and thereby maintaining the native structure of the proteins. Besides, zwitterionic polymer is electrostatically neutral and has negligible electrostatic effect on the bonded enzyme, which is conducive to the conformational stability of the immobilized proteins.
For lipase immobilization, many studies have revealed that hydrophobic supports were beneficial in protein stability and interfacial activation of lipases, while superhydrophilic zwitterionic polymers like pCBMA can stabilize proteins but are not conducive to lipase activation. In order to obtain immobilized enzymes with high activity and stability, we developed a novel series of zwitterionic polymer-grafted material by grafting zwitterionic polymers with different alkyl chains to silica nanoparticles (SNPs-pOD and SNPs-pID) (Figure 6). The polymers, pOD with long cetane side chains and pID with short alkyl side chains, are the products of the reaction of poly(maleic anhydride-alt-1-octadecene) or poly(isobutylene-alt-maleic anhydride) and N,N-dimethylethylenediamine, respectively. The zwitterionic polymers bears not only both anionic and cationic groups but also hydrophobic alkyl side chains, so lipase immobilized onto pOD- or pID-grafted SNPs (SNPs-pOD-CRL and SNPs-pID-CRL) exhibited enhanced biocatalytic activity and increased stability. Besides, comparison between the two immobilized enzymes suggests that a zwitterionic polymer with long alkyl side chains favors lipase activation rather than enzyme stability, while that with short alkyl side chains are beneficial to thermal stabilization. Then, cysteine-modified poly(glycidyl methacrylate) grafted onto silica nanoparticles and zwitterionic polymer (pOD)-coated porous poly(vinyl acetate–divinyl benzene) microspheres are designed to immobilize lipase. High performance of immobilized enzymes indicates the advantages of these zwitterionic nanoparticles for lipase immobilization and their potential for further development in various enzyme immobilizations.
|
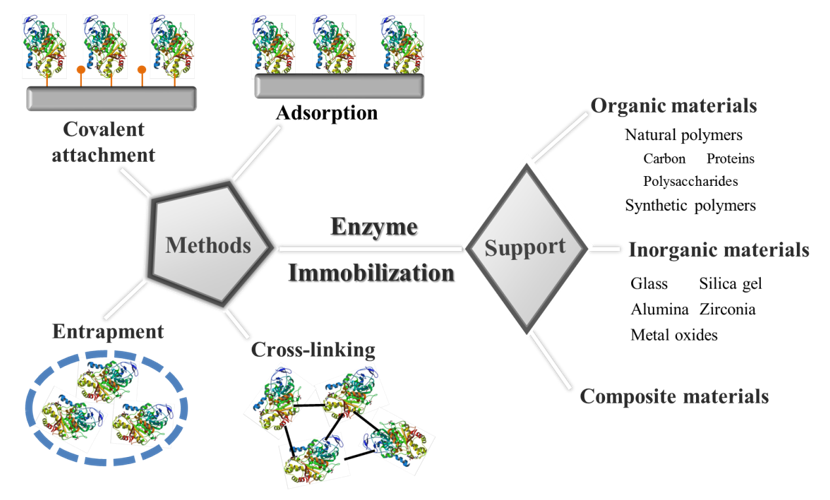 |
Figure 5. A schematic illustration of support materials and methods for enzyme immobilization. |
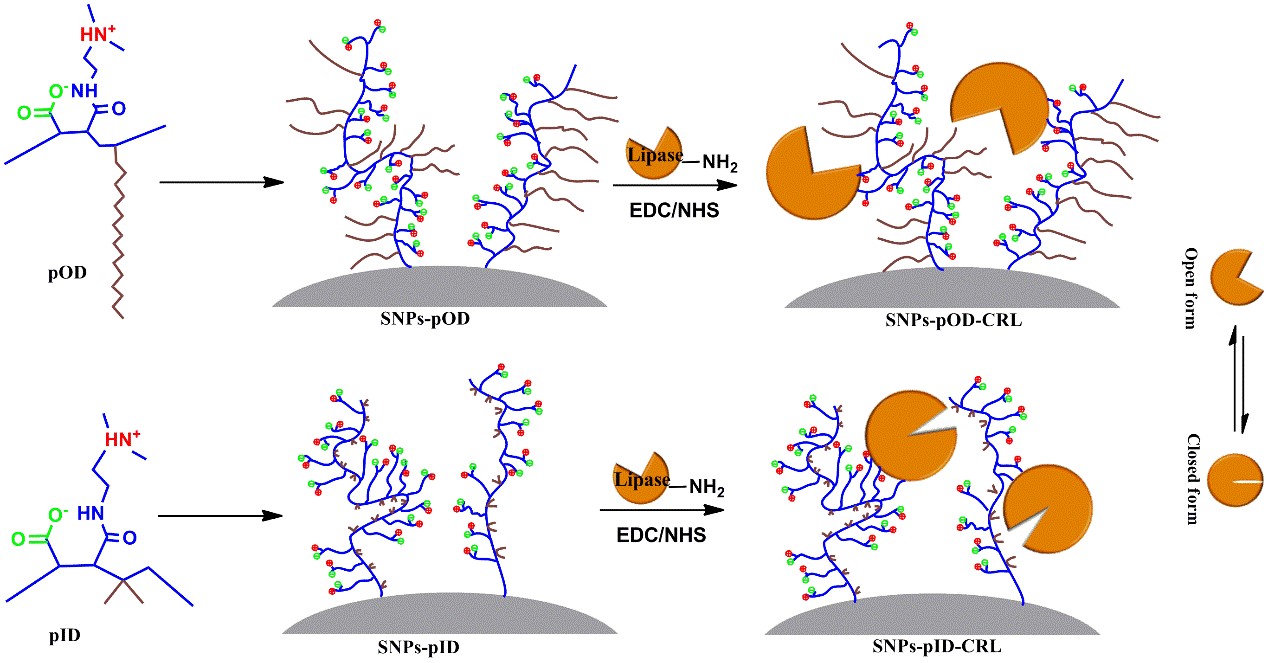 |
Figure 6. Schematic for the preparation of zwitterionic polymer-grafting onto the support and lipase immobilization. |
| |
| Back |







